Part of a series on the Hallmarks of Aging.
Proteins don’t do everything in your body, but it’s fair to say they control everything. What they don’t do directly, they catalyze. Proteins handle everything from copying DNA, to stabilizing a neuron’s physical structure, to turning starch into sugar. If something goes wrong in the body, a protein is probably to blame.
The condition where your body has a stable, healthy set of proteins–known as proteostasis–is maintained by machinery known as the “proteostasis network” (PN), which itself is composed mainly of proteins. When it fails, you may have too many or too few of a protein, or misshapen proteins that can’t do their jobs–failures which are well-known features of serious diseases like Alzheimer’s and Mad Cow disease. Because protein errors accumulate over time, impaired proteostasis is also thought to be a major contributor to certain diseases of aging.
Back to basics: How do you make a protein?
The proteostasis network starts with a ribosome translating messenger RNA into a protein.1 The RNA sequence determines the sequence of amino acids bound together linearly, known as the primary structure of the protein.
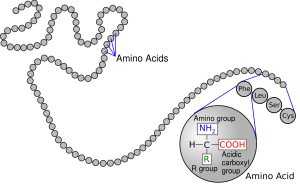
Primary protein structure. Source
From this state, the chain will naturally fold and twist into secondary structures that take the least energy to hold in place, determined by the non-covalent interactions of the different amino acids. The three most common of these are α-helixes, β-sheets and turns.
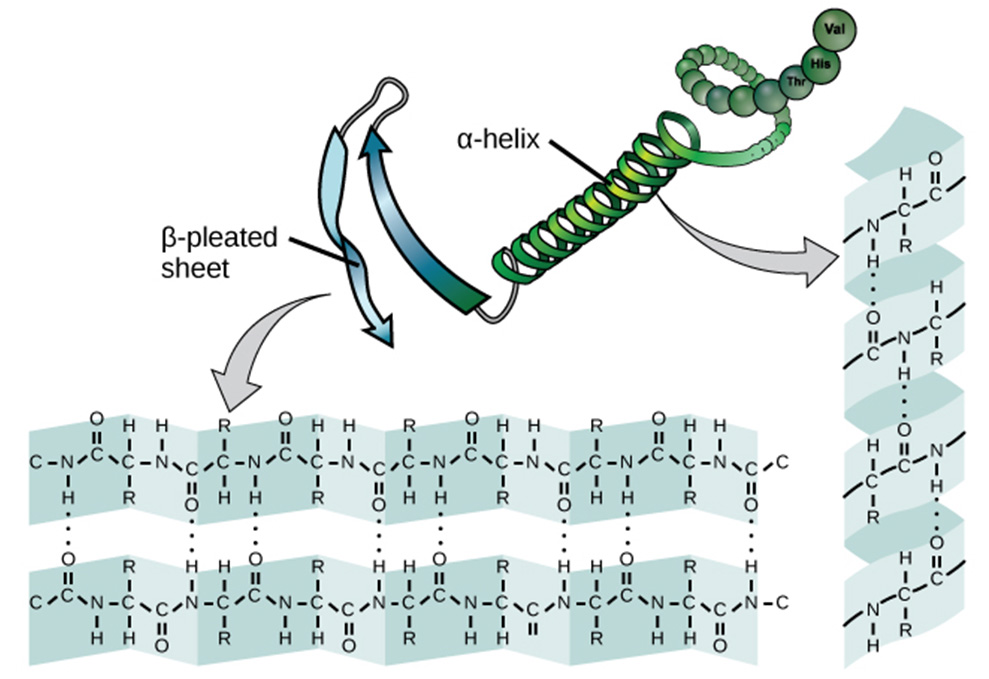
Secondary protein structure. Source
These secondary structures interact with each other much like the amino acids do, leading the polypeptide to form the three-dimensional folds of its tertiary structure. The protein may also be guided by a chaperone into a different shape that is not the global energy minimum.2 This tertiary structure may stand on its own, or join with other polypeptides. For example, hemoglobin is made of four polypeptides of two different types. The form these polypeptides take together is the protein’s quaternary structure.
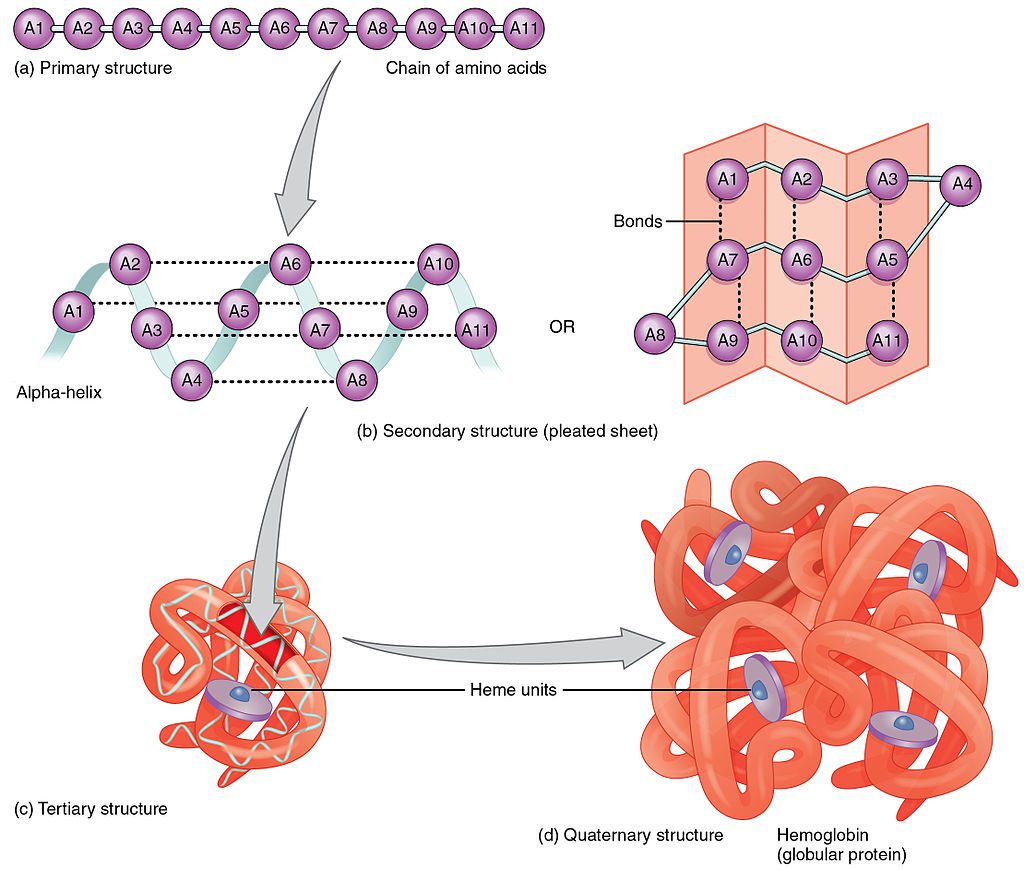
The primary, secondary, tertiary and quaternary structures that make up hemoglobin. Source
The Proteostasis Network
The proteostasis network consists of:
- Ribosomes that translate RNA into proteins. This occurs at a rate slow enough to allow secondary structures to form as translation takes place.
- Chaperones that guide polypeptides into the correct tertiary and quaternary structures.
- These include the Heat Shock Proteins, which help other proteins maintain their shape in times of stress, such as low oxygen, low pH, or high heat.
- Chaperones are helped by co-chaperone molecules that do not interact directly with the target protein, but aid the chaperone in guiding the target protein to the correct form.
- Protein degradation machinery, such as:
- Lysosomes, which are essentially bags of digestive enzymes wrapped in a membrane (to prevent rampant destruction of the cell from the inside). Similar to how immune cells digest bacteria, lysosomes can swallow a protein and break it into its constituent amino enzymes. Lysosomes are particularly active under starvation conditions, when they break down protein for energy and amino acids to fuel the most important cellular functions. This is known as macroautophagy, or just autophagy.
- Ubiquitin, a medium-sized polypeptide that can be attached to any protein to mark it for regulatory action. Ubiquitins are added to proteins by a series of enzymes, the last of which is tailored to the protein in question. Adding a chain of several ubiquitins on particular spots marks a protein for degradation via the proteasome, a large protein complex that can break down proteins into their constituent amino acids. These can then be recycled into new proteins.
Components of the proteostasis network are highly conserved, meaning they vary little across a wide variety of species. This suggests that they are extremely important, and that small changes to this system can lead to large differences in functionality.3
What can go wrong?
Misfolded proteins commonly end up in aggregations4: clumps of proteins (of the same or different kind) that are ionically bound to each other rather than forming a self-contained molecule. Certain misfolded proteins, like amyloid-beta, are also thought to be toxic to cells on their own. So what creates these misfolds?
- Environmental Stress. Changes in pH or oxidation can lead proteins to carry inappropriate charges, enabling them to ionically bond to other proteins. With too high a temperature, proteins may be able to fold into higher-energy conformations or other local optimums. Extreme temperatures on either end disrupt the non-covalent interactions between amino acids that are critical for maintaining shape. Low temperature is the cause of cryglobulinemia, in which immune system proteins solidify and block blood vessels.
- The heat shock response (HSR) system is designed specifically to prevent problems arising from environmental stress (despite the name, they deal with multiple kinds of stress). Disruption of HSR is associated with Huntington’s disease.
- Mutations and translation errors. A mutation in DNA and errors occuring in transcription both lead to RNA that codes for the wrong amino acid. The same problem can arise if the RNA is correct but the wrong amino acid is added to the polypeptide. If this acid is important to maintaining the structure of the protein, the mutation can substantially weaken the protein. Cystic fibrosis is caused by a single mutation that turns mucus secretions from thin to thick, gumming up multiple organs.
- Loss of chaperones. This can arise from failure to transcribe the relevant DNA, or because the chaperones get “stuck” in protein aggregations. Chaperone sequestration is implicated in Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis.
- Loss of degradation. Ideally, bad proteins don’t stick around long because they are degraded. Aggregations make this more difficult, because they protect inner proteins from degradation. Additionally, a lack of chaperones (perhaps lost to sequestration) may prevent proteins from traveling to an area where they can encounter a proteasome.
- Contagious aggregations. Once aggregations exist, proteins that could have degraded gracefully are able to attach to the aggregation and survive. Prion protein (PrP) is an especially pernicious version of this, because an altered conformation (PrP(Sc)) can catalyze the transformation of a healthy form of PrP to PrP(Sc). This is contagious even across species, and responsible for Mad Cow disease, scrapies, and kuru5.
You may notice that many proteostasis-related diseases are neurological. This is not a coincidence; division activates protein clean-up mechanisms, and neurons almost never divide.
What can you do about it?
Dietary restriction
Reduced caloric intake is a well-known method of extending lifespan. One possible mechanism for this is that it encourages the breakdown of proteins for fuel, which clears out damaged proteins as a byproduct.
Rapamycin
One way rapamycin may extend lifespan is that its target protein (mechanistic target of rapamycin, or mTOR) upregulates autophagy, and thus recycles bad proteins before they become problematic. Rapamycin has not been tested for longevity in humans and at the typical dosing level has number of undesirable side effects, such as immunosuppression and diabetes. It’s possible that a new regulator of mTOR will be found that gives us access to the positive effects while sidestepping the negative.
Chaperone and stabilizer replacement
These therapies inject chaperones or kinetic stabilizers to help proteins assume the correct confirmation. Both types of therapies are currently undergoing trials to assess their safety and efficacy.
Proteostasis regulator supplementation
The proteostasis network is itself regulated by multiple molecules. Supplementing them could increase the cell’s ability to correctly fold and degrade its own proteins. However, these molecules and systems are involved in so many different pathways that any intervention would likely have large side effects. The most promising idea is to create new molecules that mimic regulators under some but not all conditions.
Want more? You can find a selection of articles related to this hallmark here.
Elizabeth Van Nostrand
Elizabeth is a biologist, programmer, and science writer. She writes about science, altruism, and video games at AcesoUnderGlass.com
- While DNA transcription does influence the proteome, it is not considered part of the proteostasis network.
- However, it must be a local energy minimum–otherwise the protein would immediately transition to the closest local minimum.
- Studies of the proteostasis network (outside of neurological disease) have been done primarily in the worm C. elegans and other simple organisms. Despite the associated genes being highly conserved, it is quite possible that failures of PN in C. elegans are not representative of how the human PN actually fails.
- Conventional wisdom is that all persistent misfolded proteins end up in aggregates and all aggregates are damaging. Lee et al challenge this notion, instead suggesting that aggregations are protective against some other problem, and attempts to break them up will make things worse, not better.
- Kuru was an especially hard disease to identify. Scientists ruled out bacterial and viral causes. It hit women and children of both sexes worse than men, and appeared to be inherited–except that women inherited it from their in-laws, not their biological parents. Scientists eventually determined the cause to be a contagious, misformed prion protein, passed on through the practice of eating one’s relatives after death. Because women were considered part of their husband’s families, they ate their in-laws instead of their biological parents. Women and children were more likely to become infected because they ate brain tissue, while men primarily ate muscles.